Research Overview
The mission of Applied Genomics & Cancer Therapeutics (AGCT) program is to improve the standard of care for patients with ovarian cancer through advances in targeted therapeutics, molecular diagnostics, and early detection.
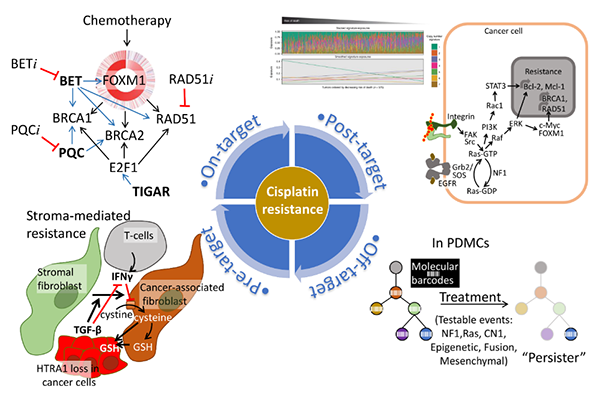 To advance targeted therapeutics and molecular diagnostics, we are currently focusing on molecular mechanisms associated with chemotherapy resistance. Given that the resistance to platinum-based chemotherapy (cisplatin and carboplatin) is a major issue in the treatment of ovarian cancer, we are investigating molecular mechanisms associated with platinum resistance. Platinum agents like cisplatin and carboplatin cause DNA damages by creating platinum-DNA adducts mainly at intra-strand GpG and ApG sites [1]. These adducts create DNA lesions and in the background of defects in the DNA repair pathway, which are a common feature in high-grade serous ovarian cancer, they result in progressive DNA damage and cell death. Consequently, cancer cells with defects in DNA repair function are selectively sensitive to these agents. Unfortunately, after several rounds of platinum-based chemotherapy, the majority of ovarian cancer acquires resistance to the treatment. Platinum resistance is multifactorial and can be conceptually organized into four categories: “pre-target”, “on-target”, “post-target”, and “off-target” mechanisms of resistance [2]. My research program explores these mechanisms by focusing on stroma-mediated resistance, DNA repair pathway, NF1/Ras pathway, and clonal selection (Figure 1). To advance targeted therapeutics and molecular diagnostics, we are currently focusing on molecular mechanisms associated with chemotherapy resistance. Given that the resistance to platinum-based chemotherapy (cisplatin and carboplatin) is a major issue in the treatment of ovarian cancer, we are investigating molecular mechanisms associated with platinum resistance. Platinum agents like cisplatin and carboplatin cause DNA damages by creating platinum-DNA adducts mainly at intra-strand GpG and ApG sites [1]. These adducts create DNA lesions and in the background of defects in the DNA repair pathway, which are a common feature in high-grade serous ovarian cancer, they result in progressive DNA damage and cell death. Consequently, cancer cells with defects in DNA repair function are selectively sensitive to these agents. Unfortunately, after several rounds of platinum-based chemotherapy, the majority of ovarian cancer acquires resistance to the treatment. Platinum resistance is multifactorial and can be conceptually organized into four categories: “pre-target”, “on-target”, “post-target”, and “off-target” mechanisms of resistance [2]. My research program explores these mechanisms by focusing on stroma-mediated resistance, DNA repair pathway, NF1/Ras pathway, and clonal selection (Figure 1). |
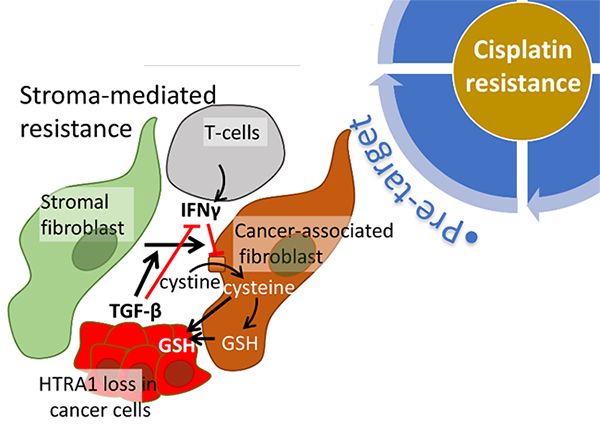 “Pre-target” resistance to platinum agents are associated with processes that act on platinum agents before they reach to the DNA. Platinum agents can react with glutathione, and the stable complex is a substrate of efflux pumps that lower intracellular accumulation of reactive platinum agents. Therefore, high levels of intracellular glutathione can lower the levels of active platinum drugs inside the cells, thereby conferring resistance in cancer cells to these agents (Figure 2). Recent studies suggest stromal factors mediate cisplatin resistance. In particular, cancer-associated fibroblasts (CAFs) can alter glutathione levels in cancer cells and confer cancer cells non-autonomous cisplatin resistance. Our ongoing project is exploring how alterations in TGF-β signaling affect CAFs, the tumor microenvironment, the levels of glutathione in cancer cells, and cisplatin resistance. We are also interested in targeting the tumor microenvironment to disrupt stroma-mediated cisplatin resistance. “Pre-target” resistance to platinum agents are associated with processes that act on platinum agents before they reach to the DNA. Platinum agents can react with glutathione, and the stable complex is a substrate of efflux pumps that lower intracellular accumulation of reactive platinum agents. Therefore, high levels of intracellular glutathione can lower the levels of active platinum drugs inside the cells, thereby conferring resistance in cancer cells to these agents (Figure 2). Recent studies suggest stromal factors mediate cisplatin resistance. In particular, cancer-associated fibroblasts (CAFs) can alter glutathione levels in cancer cells and confer cancer cells non-autonomous cisplatin resistance. Our ongoing project is exploring how alterations in TGF-β signaling affect CAFs, the tumor microenvironment, the levels of glutathione in cancer cells, and cisplatin resistance. We are also interested in targeting the tumor microenvironment to disrupt stroma-mediated cisplatin resistance.
|
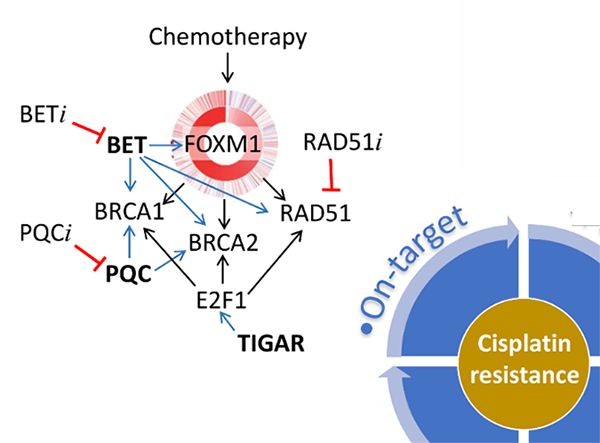 “On-target” resistance to platinum agents are associated with DNA repair processes that remove and repair platinum-DNA adducts. In particular, we are investigating an adaptive response mediated through the FOXM1 transcription factor pathway. Our previous studies reported that FOXM1 expression is deregulated in ovarian cancer [3] and is induced upon DNA damage caused by cisplatin and PARP inhibitor olaparib [4]. FOXM1 upregulates several DNA repair genes including BRCA1 and BRCA2. Consequently, the induced expression of FOXM1 by DNA-damaging chemotherapeutic agents can serve as an adaptive response to these agents and may contribute to resistance. Given that DNA repair efficiency is a strong determinant of platinum sensitivity, we are exploring different approaches to affect DNA repair efficiency in cancer cells through inhibition of protein quality control (PQC) pathway [5], transcriptional regulation on DNA repair genes by BET proteins and E2F1, and directly targeting RAD51 recombinase (Figure 3). By focusing on DNA repair genes, such as BRCA1, BRCA2, and RAD51 involved in homologous recombination (HR)-mediated DNA repairs, we aim to develop drugs that lower expression or inhibit the function of these genes and induce “BRCAness” to overcome resistance. “On-target” resistance to platinum agents are associated with DNA repair processes that remove and repair platinum-DNA adducts. In particular, we are investigating an adaptive response mediated through the FOXM1 transcription factor pathway. Our previous studies reported that FOXM1 expression is deregulated in ovarian cancer [3] and is induced upon DNA damage caused by cisplatin and PARP inhibitor olaparib [4]. FOXM1 upregulates several DNA repair genes including BRCA1 and BRCA2. Consequently, the induced expression of FOXM1 by DNA-damaging chemotherapeutic agents can serve as an adaptive response to these agents and may contribute to resistance. Given that DNA repair efficiency is a strong determinant of platinum sensitivity, we are exploring different approaches to affect DNA repair efficiency in cancer cells through inhibition of protein quality control (PQC) pathway [5], transcriptional regulation on DNA repair genes by BET proteins and E2F1, and directly targeting RAD51 recombinase (Figure 3). By focusing on DNA repair genes, such as BRCA1, BRCA2, and RAD51 involved in homologous recombination (HR)-mediated DNA repairs, we aim to develop drugs that lower expression or inhibit the function of these genes and induce “BRCAness” to overcome resistance.
|
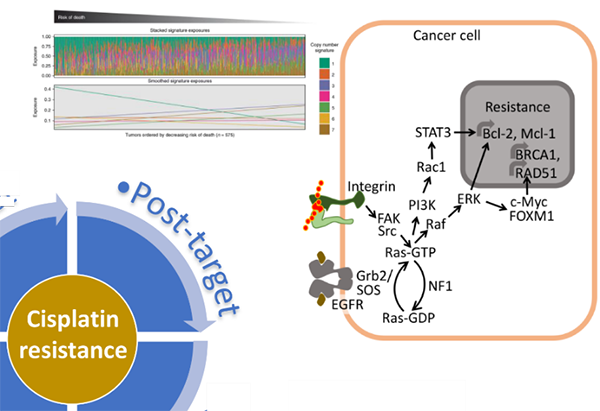 “Post-target” resistance to platinum agents are associated with downstream biological processes involving countervailing factors in pro-survival and pro-death signaling pathways induced by DNA damage response following chemotherapy. Recent evidence indicates that NF1 alterations may play an important role in modulating cancer cells’ response to chemotherapy [6, 7]. NF1 mutations are enriched in resistant or recurrent tumor samples, and copy number signature associated with NF1/Ras mutations is associated with a higher risk of death in ovarian cancer [8]. Signals transduced from NF1/Ras nexus affect pro-survival pathways as well as DNA repair pathways. We are investigating the molecular mechanisms contributing to NF1/Ras-mediated chemotherapy resistance, and we are exploring the potential clinical utility of pathway inhibitors in overcoming platinum resistance (Figure 4). “Post-target” resistance to platinum agents are associated with downstream biological processes involving countervailing factors in pro-survival and pro-death signaling pathways induced by DNA damage response following chemotherapy. Recent evidence indicates that NF1 alterations may play an important role in modulating cancer cells’ response to chemotherapy [6, 7]. NF1 mutations are enriched in resistant or recurrent tumor samples, and copy number signature associated with NF1/Ras mutations is associated with a higher risk of death in ovarian cancer [8]. Signals transduced from NF1/Ras nexus affect pro-survival pathways as well as DNA repair pathways. We are investigating the molecular mechanisms contributing to NF1/Ras-mediated chemotherapy resistance, and we are exploring the potential clinical utility of pathway inhibitors in overcoming platinum resistance (Figure 4).
|
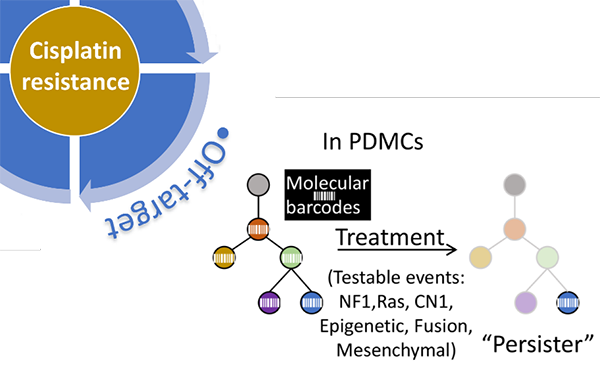 Finally, “Off-target” mechanisms of resistance are associated with alterations in gene expression and epigenetic regulations that are not directly involved in DNA damage response. We are exploring changes in gene expression, genetic and epigenetic alterations enriched in surviving “persister” cells following chemotherapy treatment. Our approach will leverage molecular barcoding, patient-derived organoid cultures, and next-generation sequencing to track clones with selective advantage upon drug treatment (Figure 5). These approaches are expected to produce insights into new “off-target” as well as other “on-target” and “post-target” mechanisms of resistance. Finally, “Off-target” mechanisms of resistance are associated with alterations in gene expression and epigenetic regulations that are not directly involved in DNA damage response. We are exploring changes in gene expression, genetic and epigenetic alterations enriched in surviving “persister” cells following chemotherapy treatment. Our approach will leverage molecular barcoding, patient-derived organoid cultures, and next-generation sequencing to track clones with selective advantage upon drug treatment (Figure 5). These approaches are expected to produce insights into new “off-target” as well as other “on-target” and “post-target” mechanisms of resistance.
|
- Jamieson, E.R. and S.J. Lippard, Structure, Recognition, and Processing of Cisplatin-DNA Adducts. Chem Rev, 1999. 99(9): p. 2467-98.
- Galluzzi, L., et al., Molecular mechanisms of cisplatin resistance. Oncogene, 2012. 31(15): p. 1869-83.
- Zhang, X., et al., Targeting of mutant p53-induced FoxM1 with thiostrepton induces cytotoxicity and enhances carboplatin sensitivity in cancer cells. Oncotarget, 2014.
- Fang, P., et al., Olaparib-induced Adaptive Response Is Disrupted by FOXM1 Targeting that Enhances Sensitivity to PARP Inhibition. Mol Cancer Res, 2018. 16(6): p. 961-973.
- Bastola, P., et al., VCP inhibitors induce endoplasmic reticulum stress, cause cell cycle arrest, trigger caspase-mediated cell death and synergistically kill ovarian cancer cells in combination with Salubrinal. Mol Oncol, 2016.
- Patch, A.M., et al., Whole-genome characterization of chemoresistant ovarian cancer. Nature, 2015. 521(7553): p. 489-94.
- Schwarz, R.F., et al., Spatial and temporal heterogeneity in high-grade serous ovarian cancer: a phylogenetic analysis. PLoS Med, 2015. 12(2): p. e1001789.
- Macintyre, G., et al., Copy number signatures and mutational processes in ovarian carcinoma. Nat Genet, 2018. 50(9): p. 1262-1270.
 To advance targeted therapeutics and molecular diagnostics, we are currently focusing on molecular mechanisms associated with chemotherapy resistance. Given that the resistance to platinum-based chemotherapy (cisplatin and carboplatin) is a major issue in the treatment of ovarian cancer, we are investigating molecular mechanisms associated with platinum resistance. Platinum agents like cisplatin and carboplatin cause DNA damages by creating platinum-DNA adducts mainly at intra-strand GpG and ApG sites [1]. These adducts create DNA lesions and in the background of defects in the DNA repair pathway, which are a common feature in high-grade serous ovarian cancer, they result in progressive DNA damage and cell death. Consequently, cancer cells with defects in DNA repair function are selectively sensitive to these agents. Unfortunately, after several rounds of platinum-based chemotherapy, the majority of ovarian cancer acquires resistance to the treatment. Platinum resistance is multifactorial and can be conceptually organized into four categories: “pre-target”, “on-target”, “post-target”, and “off-target” mechanisms of resistance [2]. My research program explores these mechanisms by focusing on stroma-mediated resistance, DNA repair pathway, NF1/Ras pathway, and clonal selection (Figure 1).
To advance targeted therapeutics and molecular diagnostics, we are currently focusing on molecular mechanisms associated with chemotherapy resistance. Given that the resistance to platinum-based chemotherapy (cisplatin and carboplatin) is a major issue in the treatment of ovarian cancer, we are investigating molecular mechanisms associated with platinum resistance. Platinum agents like cisplatin and carboplatin cause DNA damages by creating platinum-DNA adducts mainly at intra-strand GpG and ApG sites [1]. These adducts create DNA lesions and in the background of defects in the DNA repair pathway, which are a common feature in high-grade serous ovarian cancer, they result in progressive DNA damage and cell death. Consequently, cancer cells with defects in DNA repair function are selectively sensitive to these agents. Unfortunately, after several rounds of platinum-based chemotherapy, the majority of ovarian cancer acquires resistance to the treatment. Platinum resistance is multifactorial and can be conceptually organized into four categories: “pre-target”, “on-target”, “post-target”, and “off-target” mechanisms of resistance [2]. My research program explores these mechanisms by focusing on stroma-mediated resistance, DNA repair pathway, NF1/Ras pathway, and clonal selection (Figure 1).  “Pre-target” resistance to platinum agents are associated with processes that act on platinum agents before they reach to the DNA. Platinum agents can react with glutathione, and the stable complex is a substrate of efflux pumps that lower intracellular accumulation of reactive platinum agents. Therefore, high levels of intracellular glutathione can lower the levels of active platinum drugs inside the cells, thereby conferring resistance in cancer cells to these agents (Figure 2). Recent studies suggest stromal factors mediate cisplatin resistance. In particular, cancer-associated fibroblasts (CAFs) can alter glutathione levels in cancer cells and confer cancer cells non-autonomous cisplatin resistance. Our ongoing project is exploring how alterations in TGF-β signaling affect CAFs, the tumor microenvironment, the levels of glutathione in cancer cells, and cisplatin resistance. We are also interested in targeting the tumor microenvironment to disrupt stroma-mediated cisplatin resistance.
“Pre-target” resistance to platinum agents are associated with processes that act on platinum agents before they reach to the DNA. Platinum agents can react with glutathione, and the stable complex is a substrate of efflux pumps that lower intracellular accumulation of reactive platinum agents. Therefore, high levels of intracellular glutathione can lower the levels of active platinum drugs inside the cells, thereby conferring resistance in cancer cells to these agents (Figure 2). Recent studies suggest stromal factors mediate cisplatin resistance. In particular, cancer-associated fibroblasts (CAFs) can alter glutathione levels in cancer cells and confer cancer cells non-autonomous cisplatin resistance. Our ongoing project is exploring how alterations in TGF-β signaling affect CAFs, the tumor microenvironment, the levels of glutathione in cancer cells, and cisplatin resistance. We are also interested in targeting the tumor microenvironment to disrupt stroma-mediated cisplatin resistance. “On-target” resistance to platinum agents are associated with DNA repair processes that remove and repair platinum-DNA adducts. In particular, we are investigating an adaptive response mediated through the FOXM1 transcription factor pathway. Our previous studies reported that FOXM1 expression is deregulated in ovarian cancer [3] and is induced upon DNA damage caused by cisplatin and PARP inhibitor olaparib [4]. FOXM1 upregulates several DNA repair genes including BRCA1 and BRCA2. Consequently, the induced expression of FOXM1 by DNA-damaging chemotherapeutic agents can serve as an adaptive response to these agents and may contribute to resistance. Given that DNA repair efficiency is a strong determinant of platinum sensitivity, we are exploring different approaches to affect DNA repair efficiency in cancer cells through inhibition of protein quality control (PQC) pathway [5], transcriptional regulation on DNA repair genes by BET proteins and E2F1, and directly targeting RAD51 recombinase (Figure 3). By focusing on DNA repair genes, such as BRCA1, BRCA2, and RAD51 involved in homologous recombination (HR)-mediated DNA repairs, we aim to develop drugs that lower expression or inhibit the function of these genes and induce “BRCAness” to overcome resistance.
“On-target” resistance to platinum agents are associated with DNA repair processes that remove and repair platinum-DNA adducts. In particular, we are investigating an adaptive response mediated through the FOXM1 transcription factor pathway. Our previous studies reported that FOXM1 expression is deregulated in ovarian cancer [3] and is induced upon DNA damage caused by cisplatin and PARP inhibitor olaparib [4]. FOXM1 upregulates several DNA repair genes including BRCA1 and BRCA2. Consequently, the induced expression of FOXM1 by DNA-damaging chemotherapeutic agents can serve as an adaptive response to these agents and may contribute to resistance. Given that DNA repair efficiency is a strong determinant of platinum sensitivity, we are exploring different approaches to affect DNA repair efficiency in cancer cells through inhibition of protein quality control (PQC) pathway [5], transcriptional regulation on DNA repair genes by BET proteins and E2F1, and directly targeting RAD51 recombinase (Figure 3). By focusing on DNA repair genes, such as BRCA1, BRCA2, and RAD51 involved in homologous recombination (HR)-mediated DNA repairs, we aim to develop drugs that lower expression or inhibit the function of these genes and induce “BRCAness” to overcome resistance. “Post-target” resistance to platinum agents are associated with downstream biological processes involving countervailing factors in pro-survival and pro-death signaling pathways induced by DNA damage response following chemotherapy. Recent evidence indicates that NF1 alterations may play an important role in modulating cancer cells’ response to chemotherapy [6, 7]. NF1 mutations are enriched in resistant or recurrent tumor samples, and copy number signature associated with NF1/Ras mutations is associated with a higher risk of death in ovarian cancer [8]. Signals transduced from NF1/Ras nexus affect pro-survival pathways as well as DNA repair pathways. We are investigating the molecular mechanisms contributing to NF1/Ras-mediated chemotherapy resistance, and we are exploring the potential clinical utility of pathway inhibitors in overcoming platinum resistance (Figure 4).
“Post-target” resistance to platinum agents are associated with downstream biological processes involving countervailing factors in pro-survival and pro-death signaling pathways induced by DNA damage response following chemotherapy. Recent evidence indicates that NF1 alterations may play an important role in modulating cancer cells’ response to chemotherapy [6, 7]. NF1 mutations are enriched in resistant or recurrent tumor samples, and copy number signature associated with NF1/Ras mutations is associated with a higher risk of death in ovarian cancer [8]. Signals transduced from NF1/Ras nexus affect pro-survival pathways as well as DNA repair pathways. We are investigating the molecular mechanisms contributing to NF1/Ras-mediated chemotherapy resistance, and we are exploring the potential clinical utility of pathway inhibitors in overcoming platinum resistance (Figure 4). Finally, “Off-target” mechanisms of resistance are associated with alterations in gene expression and epigenetic regulations that are not directly involved in DNA damage response. We are exploring changes in gene expression, genetic and epigenetic alterations enriched in surviving “persister” cells following chemotherapy treatment. Our approach will leverage molecular barcoding, patient-derived organoid cultures, and next-generation sequencing to track clones with selective advantage upon drug treatment (Figure 5). These approaches are expected to produce insights into new “off-target” as well as other “on-target” and “post-target” mechanisms of resistance.
Finally, “Off-target” mechanisms of resistance are associated with alterations in gene expression and epigenetic regulations that are not directly involved in DNA damage response. We are exploring changes in gene expression, genetic and epigenetic alterations enriched in surviving “persister” cells following chemotherapy treatment. Our approach will leverage molecular barcoding, patient-derived organoid cultures, and next-generation sequencing to track clones with selective advantage upon drug treatment (Figure 5). These approaches are expected to produce insights into new “off-target” as well as other “on-target” and “post-target” mechanisms of resistance.